Summer Project 2: Das Lab
One of the most exciting breakthroughs in structural biology and machine learning was AlphaFold’s performance at the CASP14 competition - proving the superiority of deep neural network models in protein structure prediction. More recently, AlphaFold 3 has expanded to include the prediction of biomolecular interactions and nucleic acids. However, there remain noticeable limitations in predicting large, RNA only complexes as some AlphaFold3 models contain severe steric clashes and breaks in phosphodiester backbone (3). Most attribute the poor performance of AF3 on RNA structure prediction to the deficiency of RNA structural data in the pdb. As a result, many have suggested augmenting the sparse 3D RNA structural data with secondary structure chemical mapping data. The idea was to train a deep neural network to predict, given an RNA sequence, its corresponding chemical mapping reactivity profile. This model could then be finetuned on downstream tasks on smaller datasets- including tertiary structure prediction. The first startup company Atomic-AI released a publication of its model ATOM-1 (not available to the public) demonstrating the feasability of this idea. Similarly, the Das Lab released a publication describing the model RibonanzaNet trained on millions of RNA structure’s chemical mapping data that similarly performs well on downstream tasks. This model designed by Shujun-He with insight from various other Kaggle models is free to use and available here
A few years ago, the Das Lab also released a paper with benchmark datasets of RNA secondary structure, chemical mapping profiles, and ligand binding equilibrium constants. Additionally, this paper released a statistical model, EternaFold, that used the baseline CONTRAfold architecture but trained on all three unique datasets. EternaFold was shown to outperform all current models on all prediction tasks.
Using the EternaFold dataset, I evaluated RibonanzaNet’s performance on all tasks, regenerating figures in the EternaBench paper as well as creating my own based on raw data. I returned to this task a few months later, also adding RNA-FM into the mix.
RibonanzaNet Secondary Structure Evaluation:
Regenerated heatmap of Z-scores of F-scores from EternaBench paper on ArchiveII dataset of secondary structures:

This is RibonanzaNet-SS trained with different checkpoints that vary in magnitude by the amount of training data. RibonanaNet is the RibonanzaNet-SS model trained on all 2 million sequences in the Ribonanza Dataset. RibonanzaNet A, B, C, D are models trained on 140k, 14k, 7k, and 1.4k training data, respectively.
RibonanzaNet Chemical Mapping Evaluation
A dataset of chemical mapping profiles using the SHAPE reagent 1M7 of natural and human-designed RNAs was compiled, and correlation coefficients were computed, with z-score heatmaps generated.
Here is a barchart comparing Pearson Correlation Coefficients with RibonanzaNet-SS, RibonanzaNet-2A3, and RibonanzaNet-DMS.
However, I am not confident that this 1M7 is a good dataset to evaluate performance of models, considering it is very noisy and spiky. Here are two reactivity profiles that show 1M7 raw data with some of the package predicted reactivity profiles. Note, the simulated reactivity profile form secondary structure prediction models comes from converting base pairing matrices to probability unpaired vectors (punp vectors) which is outlined in the Das Lab arnie wrapper.
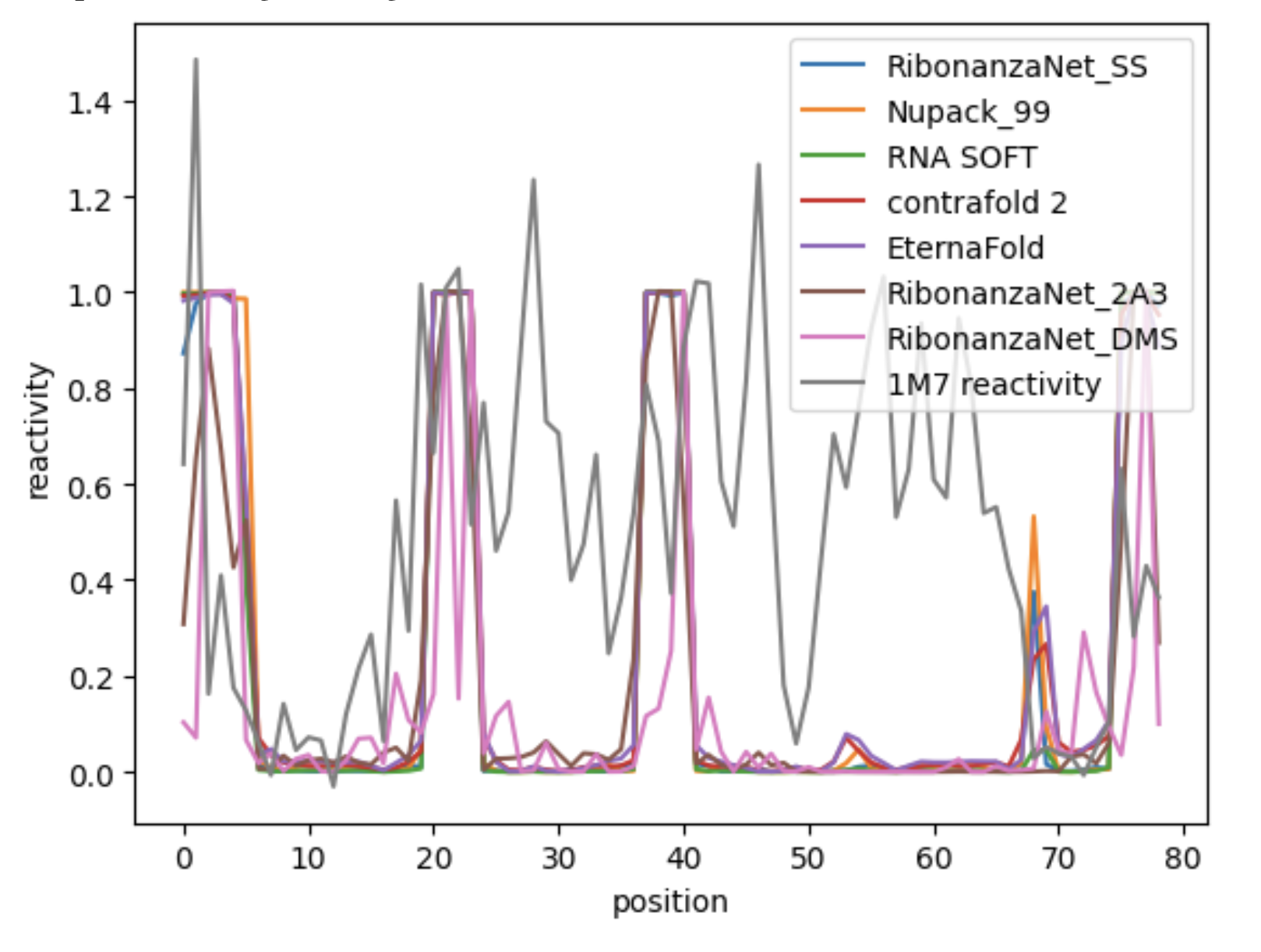

You can play around with reactivity profiles yourself my copying the folder in this repository /notebooks/eternabench_chemmapping_simulation (and all the contents within it) here :
RibonanzaNet Riboswitch Ligand-Binding Prediction Evaluation
The last metric evaluated was the preditive ability of each model to predict the ligand-binding affinity of riboswitches indirectly through the probability of the the terminal base pair in the aptamer hairpin. The predicted binding affinity based on constrained partition function calcuations will also be evaluated in my next project that includes how I finetuned RibonanzaNet on this data.
These riboswitches were designed so that upon binding one of the input ligand aptamers, flavin mononucleotide (FMN), theophylline (Theo), or tryptophan (Trp), the MS2 protein aptamer becomes more accessible, thereby increasing binding affinity of fluorescent protein. Experimental measurements of MS2 protein binding affinities with and without ligand present (K+lig and K-lig) were performed on this diverse dataset.
Because the MS2 aptamer is a hairpin, a zeroshot evaluation on riboswitch binding affinities without ligand bound (K-lig) was performed using RibonanzaNet-SS bpp matrices on the EternaBench holdout dataset. Evaluating on checkpoints of RibonanzaNet-SS trained with a variety of data scales allowed us to further determine the extent that data quantity, rather than model design, improved accuracy of predictions.
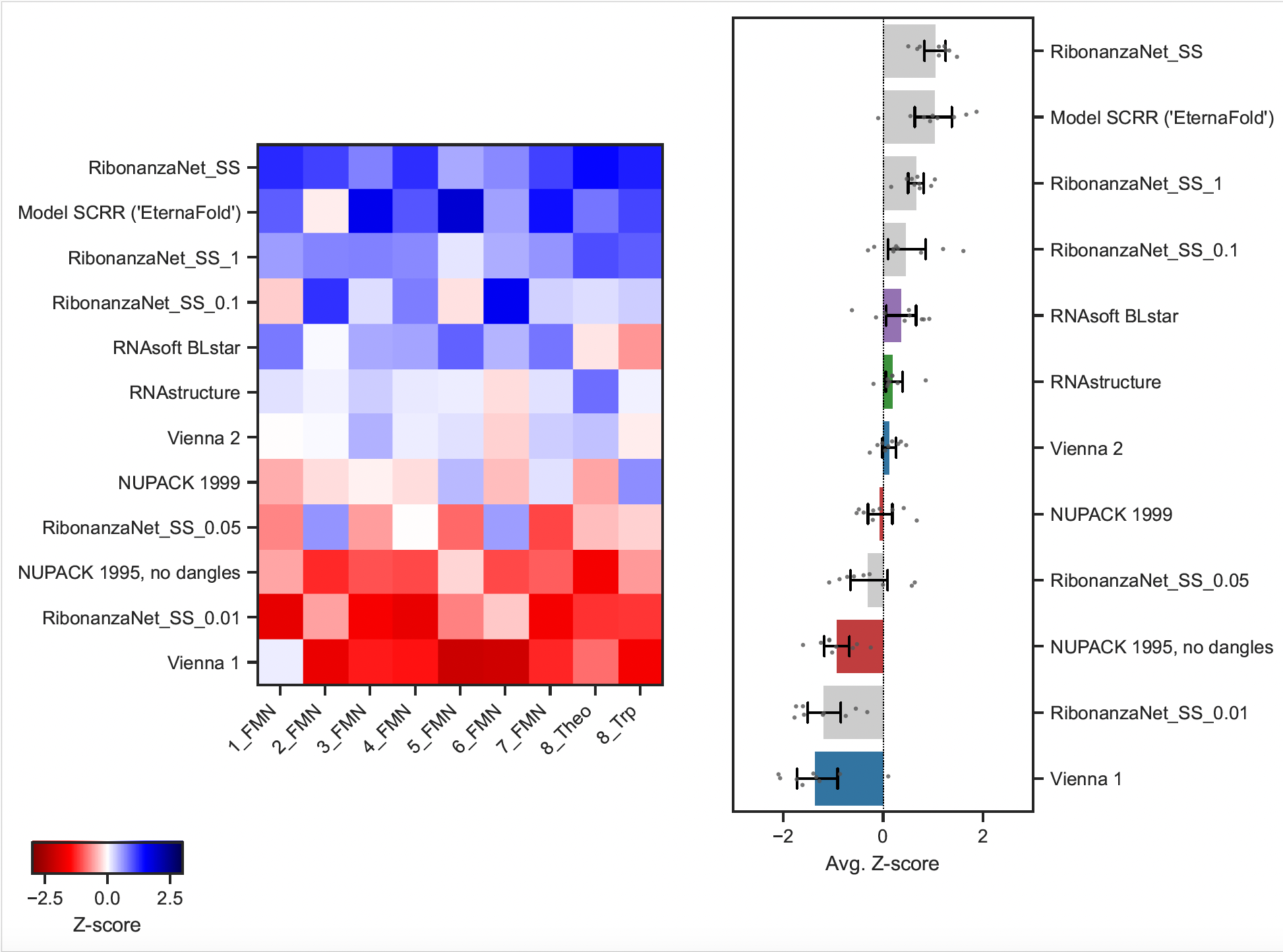
How I generated these predictions can be accessed in this github repo.
Relevant Papers:
(1) He, S.; Huang, R.; Townley, J.; Kretsch, R. C.; Karagianes, T. G.; Cox, D. B. T.; Blair, H.; Penzar, D.; Vyaltsev, V.; Aristova, E.; Zinkevich, A.; Bakulin, A.; Sohn, H.; Krstevski, D.; Fukui, T.; Tatematsu, F.; Uchida, Y.; Jang, D.; Lee, J. S.; Shieh, R.; Ma, T.; Martynov, E.; Shugaev, M. V.; Bukhari, H. S. T.; Fujikawa, K.; Onodera, K.; Henkel, C.; Ron, S.; Romano, J.; Nicol, J. J.; Nye, G. P.; Wu, Y.; Choe, C.; Reade, W.; Eterna participants; Das, R. Ribonanza: Deep Learning of RNA Structure through Dual Crowdsourcing. bioRxivorg 2024. https://doi.org/10.1101/2024.02.24.581671.
(2) Wayment-Steele, H. K.; Kladwang, W.; Strom, A. I.; Lee, J.; Treuille, A.; Becka, A.; Participants, E.; Das, R. RNA Secondary Structure Packages Evaluated and Improved by High-Throughput Experiments. Nat. Methods 2022, 19 (10), 1234–1242. https://doi.org/10.1038/s41592-022-01605-0.
(3) McDonnell, R. T.; Henderson, A. N.; Elcock, A. H. Structure Prediction of Large RNAs with AlphaFold3 Highlights Its Capabilities and Limitations. J. Mol. Biol. 2024, 436 (22), 168816. https://doi.org/10.1016/j.jmb.2024.168816.

