Rotation Project 2: Das Lab
A hypothetical scenario: the world was flash frozen in an instant by an alien species. In order to study the most dominant earthy organism - humans, the alien species decides that the primary function of humans can be deciphered by figuring out the most populated state and lowest energy state (according to the laws of thermodynamics). Thus, the aliens scientifcally conclude that the primary function of humans is to lay flat on a surface, with eyes closed, in a dreamlike state.
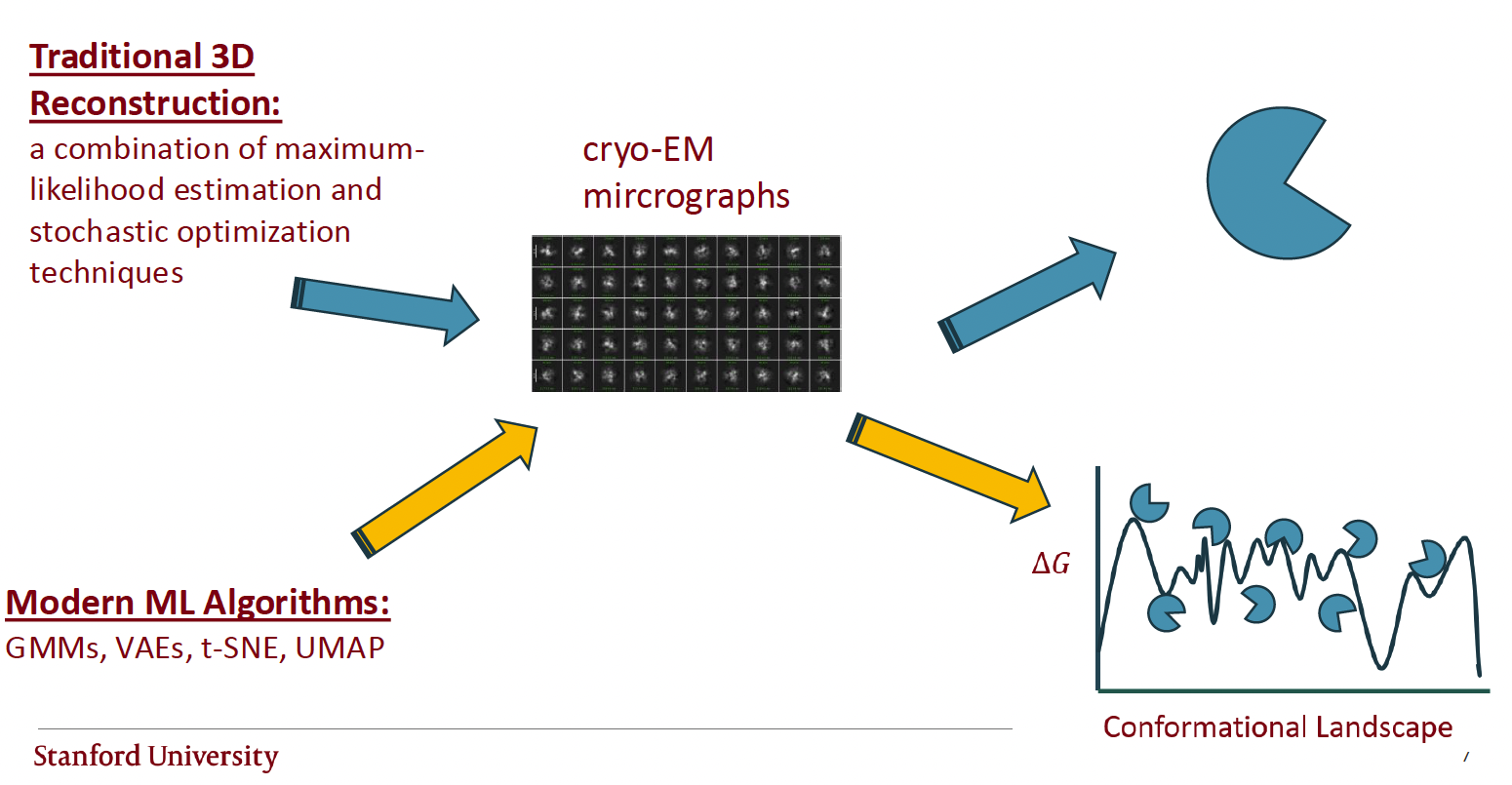
Now, this hypothetical scenario may seem ridiculous, but it is the primary and best mechanism for understanding structural biology at the moment. We flash freeze biological macromolecules, or use physics-based simulations, to uncover their functions based on their lowest energy or most populated structural states. However, maybe the most important function of biological macromolecules, especially RNA, is related to a state(s) corresponding with a low statistical weight?
I think it is an important goal of academic labs to develop methods for uncovering these rare states. One such means of doing this is to parse out hetereogenous ensembles of structures from cryo-EM micrographs.
In my first rotation project, I evaluated a the novel cryo-EM heterogeneity VAE, DynaMight, on simulated micrographs from an MD simulation on the Tetrahymena ribozyme. This model has shown excellent results on protein and protein complexes, however, it has not yet been evaluated on RNA-only complexes.
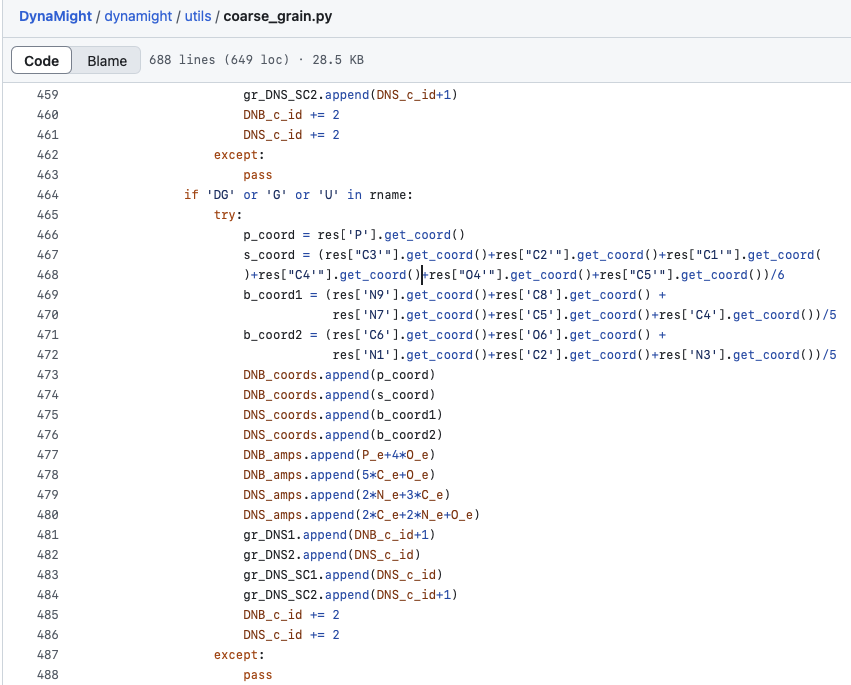
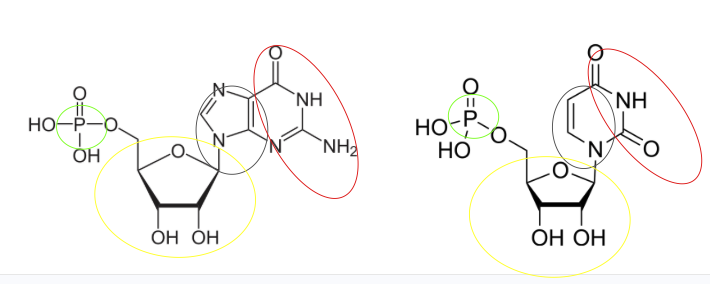
This model was not easy for me to work with which allowed me to dig deep into the underlying code for debugging. (Not to criticize the model or the code- I have never been able develop such an algorithm from scratch, what an accomplishment!). One of the most suprising finds during the debugging process came from realizing that the coarse graining script kept omitting uracils. Fortunately, at the time I was taking Carolyn Bertozzi’s Therapeutic Chemistry course. In the course she encouraged all graduate students to regularly memorize the structure of each nucleic acid. I listened and made an effort last spring to regularly draw each of the nucelobases. From this I could easily catch an error in DynaMight’s coarse graining code: guanine and uracil were given the same atomic labels, even though uracil is a completely different structure, lacking the purine-specific atoms used to coarse grain guanine.
 ”
”  ”
”
Here are the slides of my second rotation project at Stanford, in the Das Lab.
